Focus
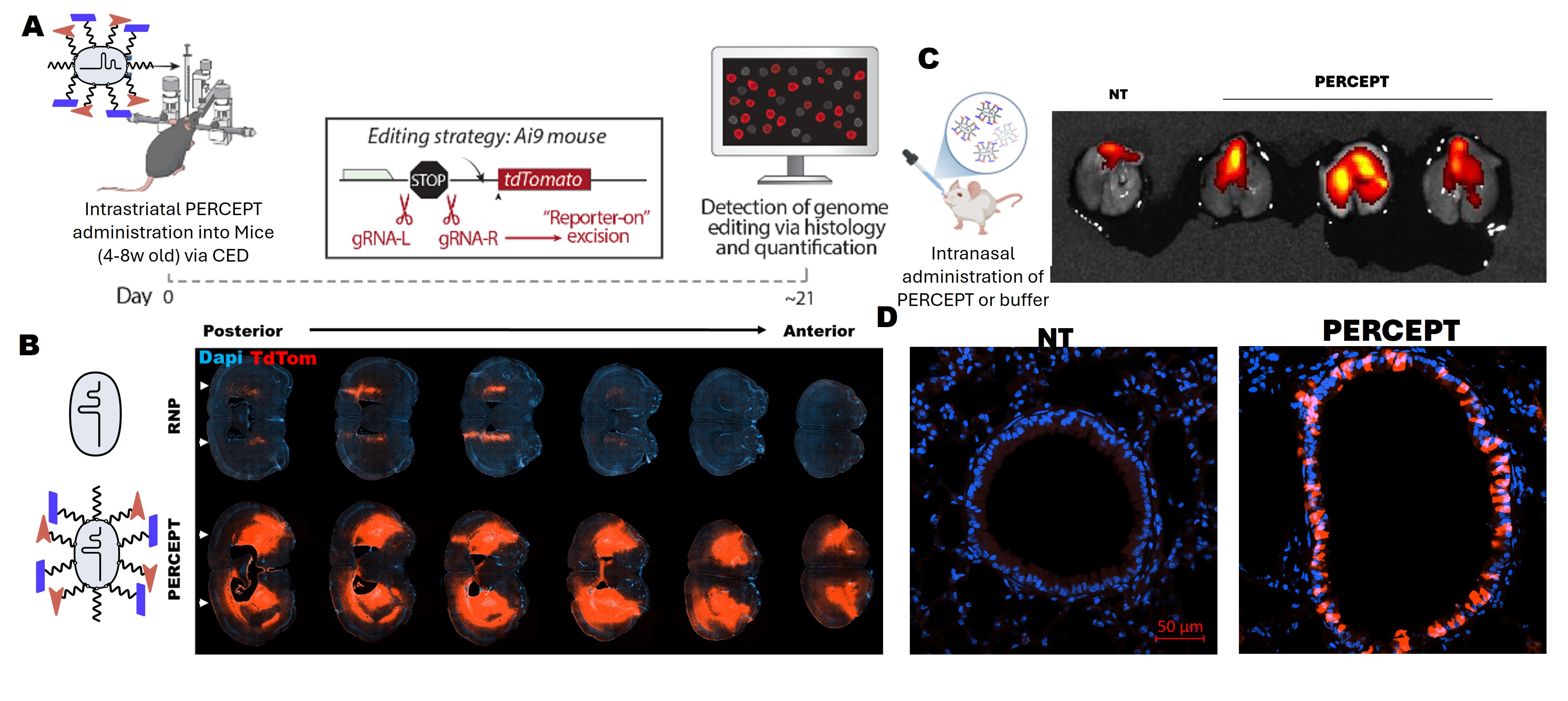
The limiting step for in vivo genome editing is delivery: getting an editor into the right cells at useful levels with controlled exposure. I develop non-viral CRISPR RNP delivery built around PERCEPT, a modular platform that allows targeting ligands and formulation chemistry be changed at the final step, enabling direct, matched in vivo comparisons. After local administration, PERCEPT has produced strong editing in the CNS and retina and has also performed well in lung and skeletal muscle, motivating a single platform to iterate across tissue environments and pinpoint the constraints that limit editing.